DNA Recognition Modules
A-3. Search for Novel DNA Recognition Protein Modules
N. Yachie, Univ. Tokyo
yachiesynbiol.rcast.u-tokyo.ac.jp
Overview
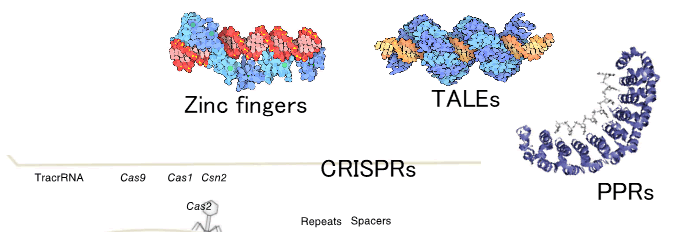
The genome-editing tools that have been practically implemented to date are based on naturally occurring periodic sequence repeats.
Software capable of quickly detecting periodic repeats in a large-scale genetic database was developed and used to identify a group of candidate modules.
Achievements
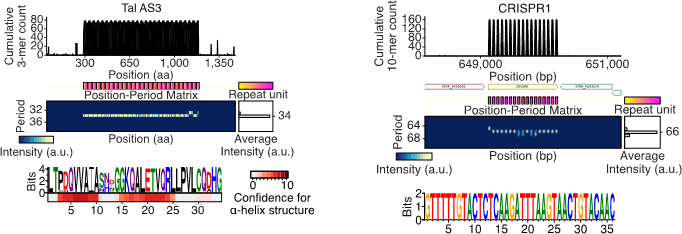
- The SPADE software package has been released.
- The SPADE software accurately identifies the repeat sequences recognized by CRISPR, Zinc finger, TALE, PPR, and other existing editing modules.
- Its accuracy of detection is comparable to those of software used to detect the effects of the versatile and widely adopted CRISPR system.
- SPADE outperformed software used to detect common repeat sequences.
- Discovery of a cluster of 128 potential candidate genes for new genome-editing proteins.
Summary
Development of software that evaluates genomic and metagenomic resources and screens for genes for new genome-editing modules based on sequence periodicity.
Technical advantages
- Search methods that are not restricted by the type of editing tools
- High-throughput screening of large-scale genome resources
Challenges for Practical Application
- Proof-of-concept experiments to demonstrate the effects of new candidates
Resources Available
- The SPADE software package
Notes
A cluster of 128 candidate genes for new genome-editing protein modules was identified as a result of running SPADE on the whole genomes of 7006 microorganisms. SPADE detects periodic sequence repeats automatically. An automated experimental system for evaluating candidate editing modules is under construction.