Protein structure, dynamics - Biophysical methods, Bioimaging, Proteomics, Computational simulations in Molecular & Cellular Biology
Through analysis of the dynamic structural properties of proteins, we will elucidate the functional role of fluctuations in protein structures. In particular, we are conducting research focusing on the elucidation of new functions of intrinsically disordered regions (IDRs). The characteristic of the research carried out in this laboratory is to proceed with the development of original research based on the development of new structural analysis technology.

Tteam is currently involved in projects related to liquid-liquid phase separation, chirality, allostery and related biological phenomena, through a variety of experimental and computational methods.
Proteins harboring intrinsically disordered regions (IDRs) form biomolecular condensates through liquid-liquid phase separation (LLPS) in live cells.
The liquid-like granular condensates are composed of variety of proteins and RNAs. By choosing the stress granule and ALS-related cytoplasmic granules as models, our
group tries to reveal the time-dependent change in the constitution of those biomolecular condensates. The project runs in collaboration with Prof. Rhee of Seoul National University, Korea.

In collaboration with Dr. Ogura (National Institute of Advanced Industrial Science and Technology, Tsukuba), our group is tackling to image the biomolecular condensates in live cells at nano-scale resolution. The project aims to unveil how various biomolecules assemble to generate condensates.

Proteins with the intrinsically disordered regions (IDRs) mediate biomolecular granulation via liquid-liquid phase separation (LLPS). We explore the molecular mechanism of protein granulation with advanced NMR spectroscopy
combined with molecular biology and theoretical simulation like molecular dynamics simulation. The research outcomes will unveil the pathogenic mechanisms of neurodegenerative diseases, including ALS and Alzheimer’s diseases.
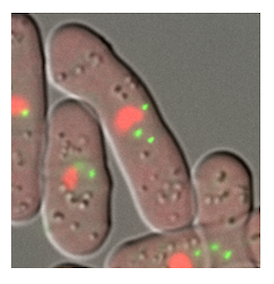
DNA is packed in the cell nucleus as chromatin fibers comprising nucleosomes (protein-DNA complex). Gene regulation in the cell nucleus
requires a dynamic structural change of chromatin. In collaboration with Prof. Awazu and Prof. Ueno in the university, our group explores how the chromatin structure dynamics engage in gene regulation with multimodal approaches using light microscopy, electron microscopy, and mathematical modeling to integrate the experimental observations.
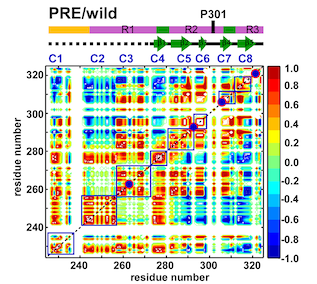
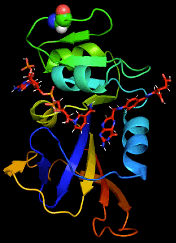
By combining Nuclear Magnetic Resonance experiments with Computational Modeling, we are exploring the mechanism of allostery in dihydrofolate reductase (DHFR) as revealed by a novel mutation. Via Molecular Dynamics (MD) simulations, we explore changes to changes in dynamics that may explain changes in catalytic activity.
Led by Prof. Shinichi Tate
Check our our latest activities!
We discuss the implications of considering the dynamics of a protein in a protein-protein complex, as modelled implicitly and explicitly with methods dependent on elastic network models. We further propose how such an explicit consideration can be applied to understand critical protein-protein contacts that can be targeted in future studies.
Read More
Fertilized eggs begin to translate mRNAs at appropriate times and placements to control development, but how the translation is regulated remains unclear. Here, we found that pou5f3 mRNA encoding a transcriptional factor essential for development formed granules in a dormant state in zebrafish oocytes...
Read More
Topic from special edition・FUS液液相分離の機能と細胞内相分離制御機構(安田 恭大・楯 真一)
Read MoreMolecular Biophysics Laboratory,
Hiroshima University,
Graduate School of Science,
Mathematical Molecular Life Science,
Faculty of Science / Department of Chemistry,
1-3-1 Kagamiyama, Higashihiroshima City,